Our World in Data Examples
Data sets
This work references data on life expectancy and causes of death from the Our World In Data organization. The life expectancy and survival rate graphics reference the specific section of the site here. The causes of death graphics reference the specific section of the site here.
life_expectancy_data <- tibble(read.csv("docs/life-expectancy.csv")) %>%
arrange(Entity, Year)
colnames(life_expectancy_data) <- c("Entity", "Code", "Year", "Expectancy")
men_survival_to_age_65 <- tibble(read.csv("docs/men-survival-to-age-65.csv")) %>%
arrange(Entity, Year)
colnames(men_survival_to_age_65) <- c("Entity", "Code", "Year", "Percent")
women_survival_to_age_65 <- tibble(read.csv("docs/women-survival-to-age-65.csv")) %>%
arrange(Entity, Year)
colnames(women_survival_to_age_65) <- c("Entity", "Code", "Year", "Percent")
causes <- c("Executions", "Meningitis", "Lower_Respiratory_Infections",
"Intestinal_Infectious_Diseases", "Protein-Energy_Malnutrition", "Terrorism",
"Cardiovascular_Diseases", "Dementias", "Kidney_Disease",
"Respiratory_Diseases", "Liver_Diseases", "Digestive_Diseases",
"Hepatitis", "Cancers", "Parkinson_Disease", "Fire", "Malaria", "Drowning",
"Homicide", "HIV/AIDS", "Drug_Use_Disorders", "Tuberculosis",
"Road_Injuries", "Maternal_Disorders", "Neonatal_Disorders", "Alcohol_Use_Disorders",
"Natural_Disasters", "Diarrheal_Diseases", "Heat_And_Cold_Exposure",
"Nutritional_Deficiencies", "Suicide", "Conflict", "Diabetes", "Poisonings")
causes_of_death <- tibble(read.csv("docs/annual-number-of-deaths-by-cause.csv")) %>%
arrange(Entity, Year)
colnames(causes_of_death) <- c("Entity", "Code", "Year", all_of(causes))
causes_of_death <- causes_of_death %>% mutate(Executions = as.numeric(Executions))Plotting functions
The code snippet here shows the plotting designs used in following graphics tabs.
# various plot designs used in the analysis
le_plot_single <- function(data) {
ggplot(data, aes(Year, Expectancy)) +
geom_line() +
labs(
x = "Year",
y = "Life Expectancy",
title = "Life expectancy",
subtitle = "1770 to 2019",
caption = "Source: Our World In Data"
) +
theme_minimal() +
scale_y_continuous(limits = c(20, 90), breaks = seq(20, 90, by = 10)) +
theme(legend.position = "none") +
scale_colour_brewer(palette = "Dark2")
}
le_plot_multi <- function(data, color_lab) {
ggplot(data, aes(Year, Expectancy, color = Entity)) +
geom_line() +
labs(
x = "Year",
y = "Life Expectancy",
title = "Life expectancy",
color = color_lab,
subtitle = "1770 to 2019",
caption = "Source: Our World In Data"
) +
theme_minimal() +
scale_y_continuous(limits = c(20, 90), breaks = seq(20, 90, by = 10)) +
theme(legend.position = "bottom") +
scale_colour_brewer(palette = "Dark2")
}
sr_plot <- function(data, title_lab, color_lab) {
ggplot(data, aes(Year, Percent, color = Entity)) +
geom_line() +
labs(title = title_lab,
color = color_lab) +
theme_minimal() +
scale_y_continuous(limits = c(20, 100),
breaks = seq(20, 100, by = 10)) +
scale_colour_brewer(palette = "Dark2")
}
cod_plot <- function(data, xlab, end_scale, increment) {
ggplot(data, aes(Count, Cause)) +
geom_col(aes(fill = Count)) +
theme_minimal() +
theme(legend.position = 'none') +
scale_fill_distiller(palette = "Dark2") +
geom_label(
aes(label = Count),
size = 3,
hjust = -0.1,
label.size = 0.1,
label.padding = unit(0.1, "lines")
) +
scale_x_continuous(limits = c(0, end_scale), breaks = seq(0, end_scale, by = increment)) +
labs(x = xlab,
y = "")
}Life Expectancy
These graphs show period life expectancy at birth: the average number of years a newborn would live if the pattern of mortality in the given year were to stay the same throughout its life.
The World
life_expectancy_data %>%
filter(Entity == "World") %>%
le_plot_single()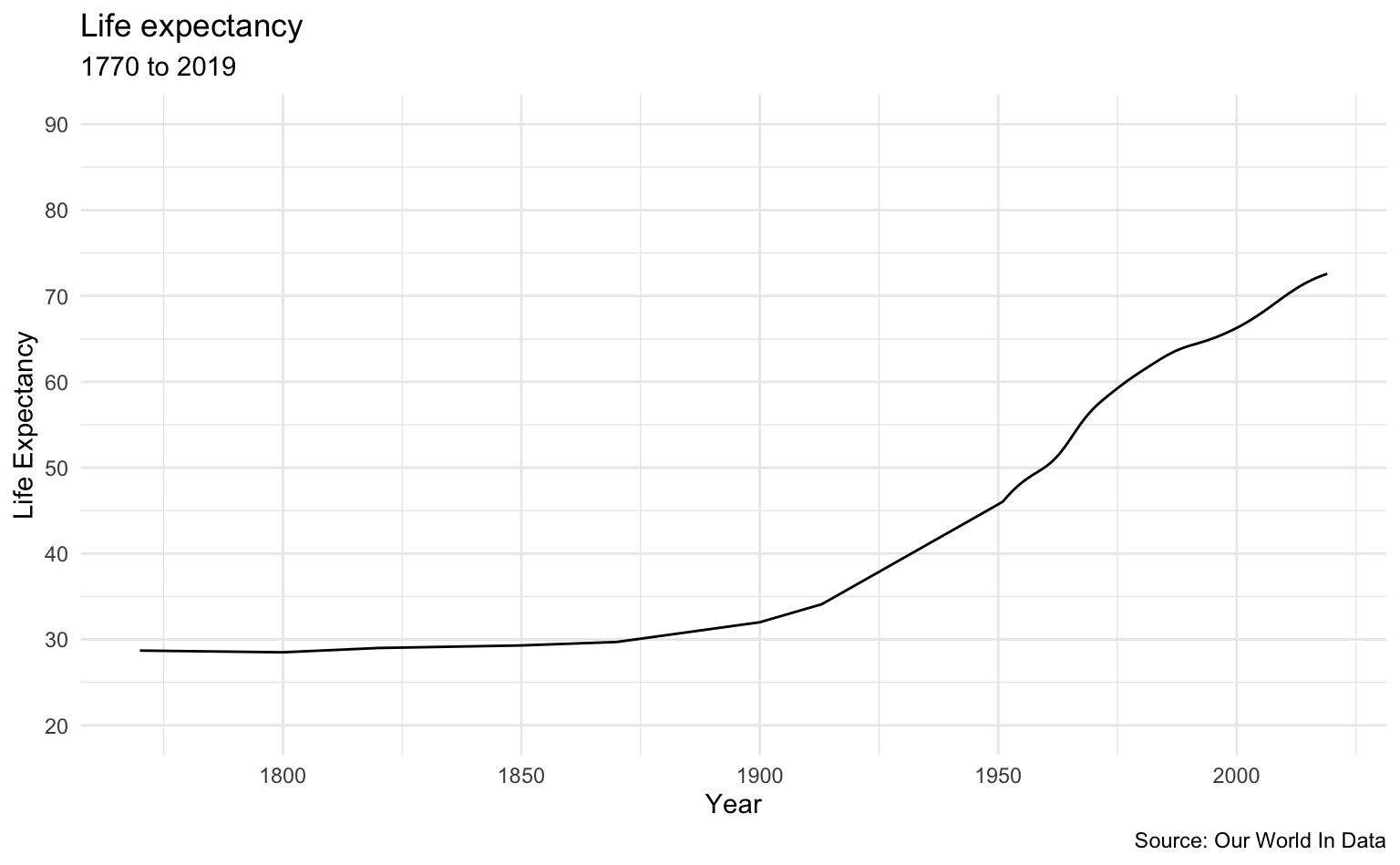
Regions
sample_list <- c("Oceania", "Europe", "Americas", "Asia", "Africa", "World")
life_expectancy_data %>%
filter(Entity %in% sample_list) %>%
le_plot_multi("Region")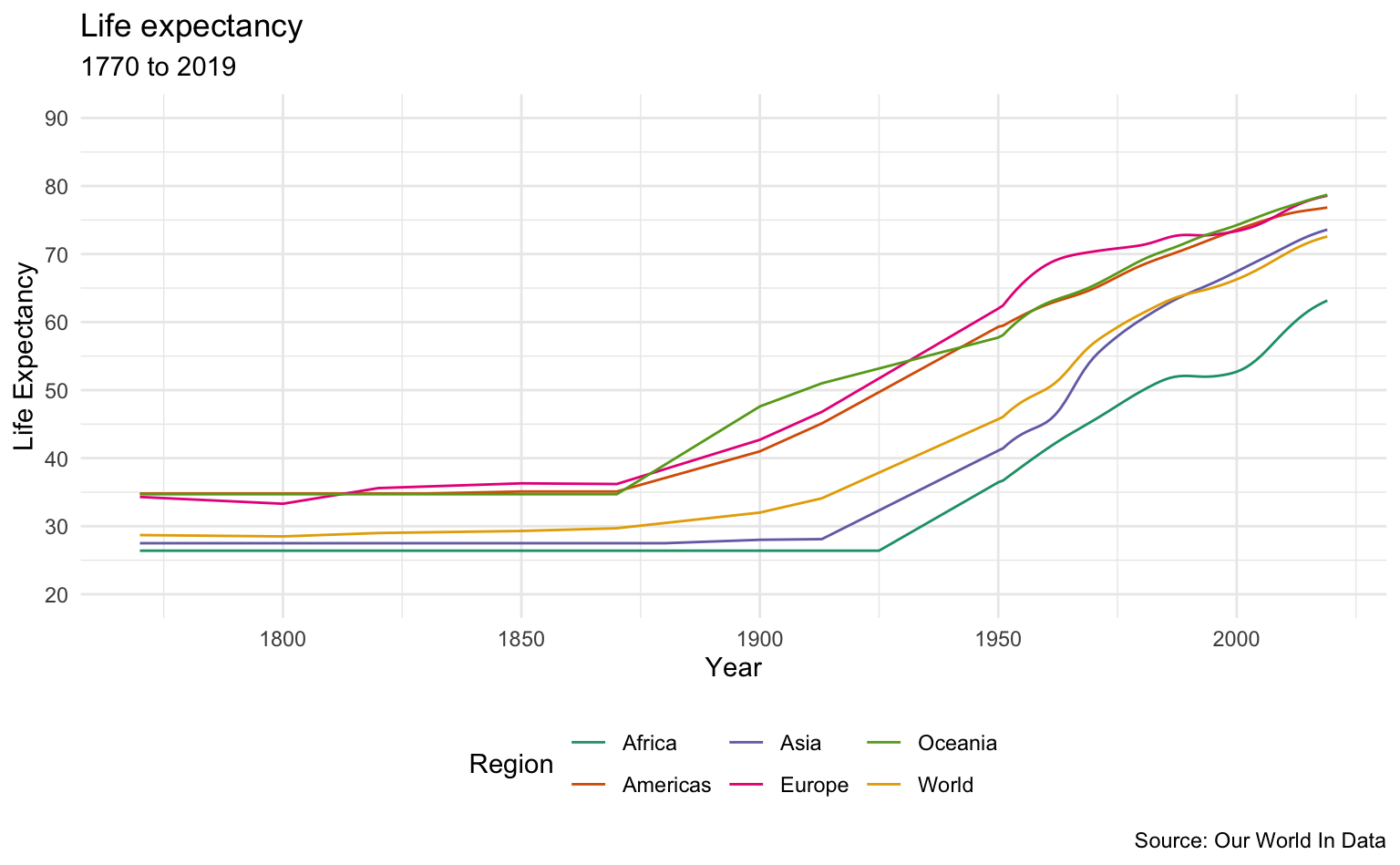
Selected Countries
sample_list <- c("Japan", "South Korea", "United Kingdom", "India", "Ethiopia", "South Africa", "United States", "World")
life_expectancy_data %>%
filter(Entity %in% sample_list) %>%
filter(Year >= 1865) %>%
le_plot_multi("Country")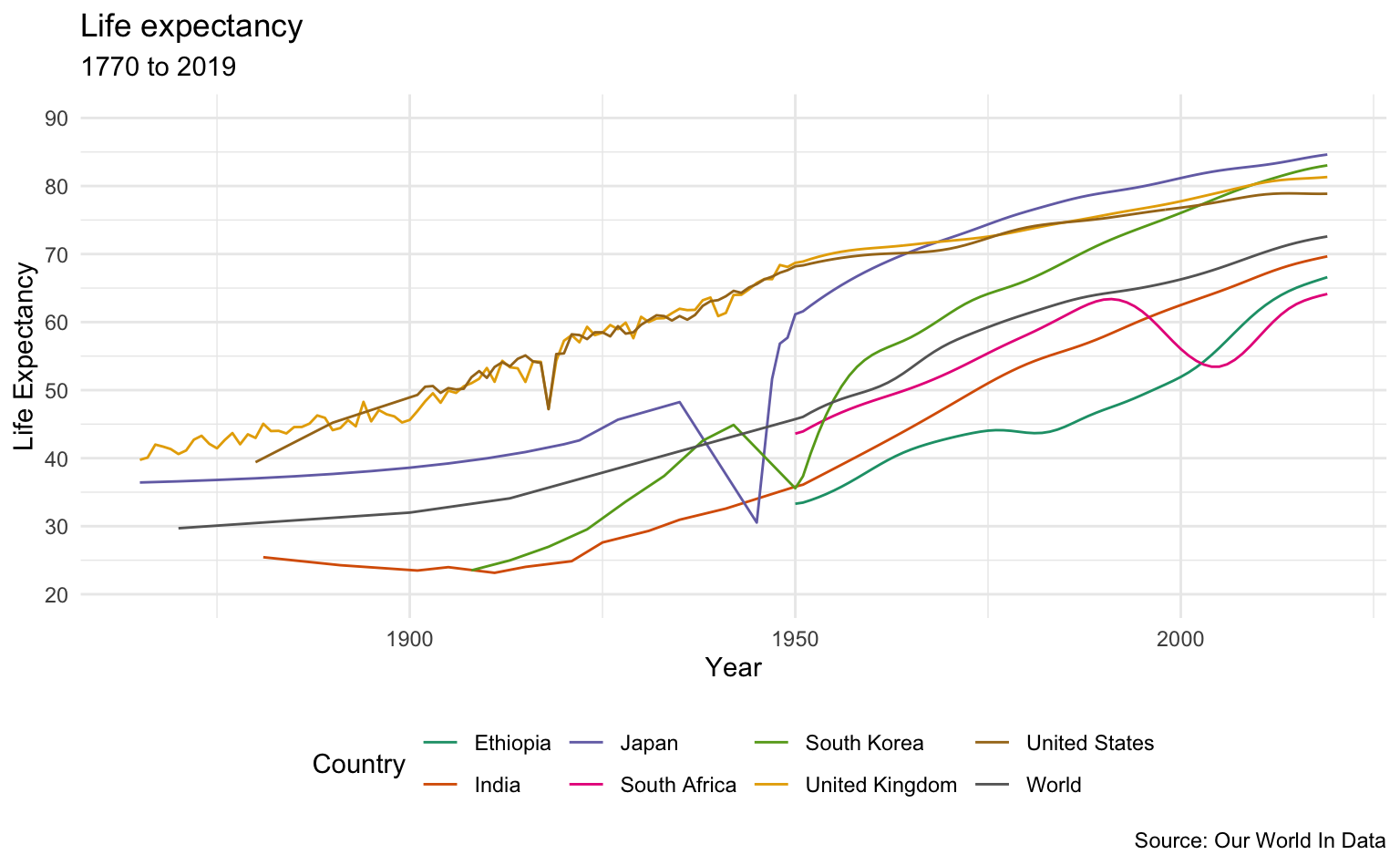
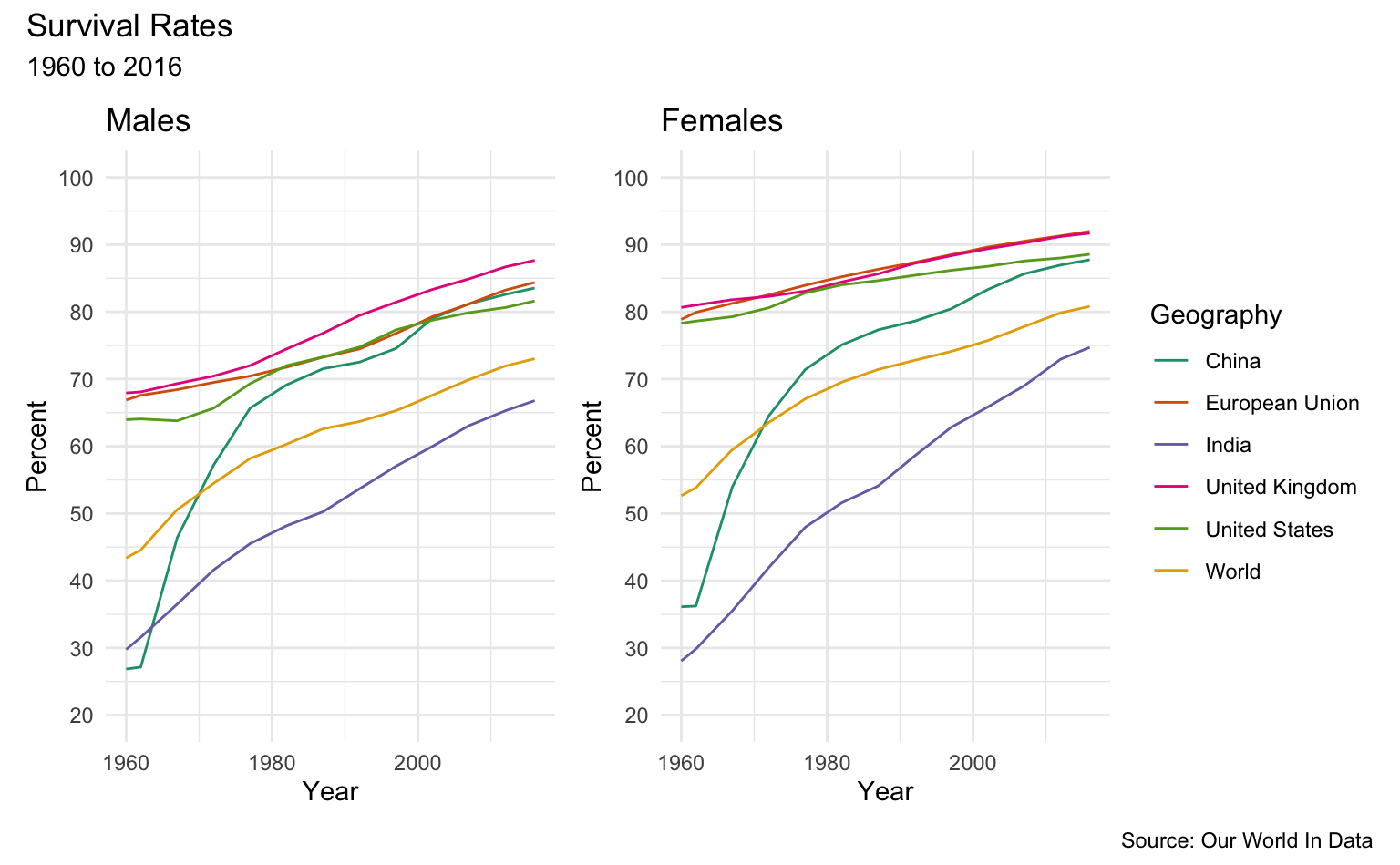
Survival Rates
These graphs show the share of the population that is expected to survive to the age of 65.
Selected Geographies
sample_list <- c("United States", "China", "Philipines", "United Kingdom", "European Union", "India", "World")
m <- men_survival_to_age_65 %>%
filter(Entity %in% sample_list) %>%
sr_plot("Males", "Geography")
w <- women_survival_to_age_65 %>%
filter(Entity %in% sample_list) %>%
sr_plot("Females", "Geography")
m + w + plot_layout(ncol = 2, guides = "collect") +
plot_annotation(
title = "Survival Rates",
subtitle = "1960 to 2016",
caption = "Source: Our World In Data"
)
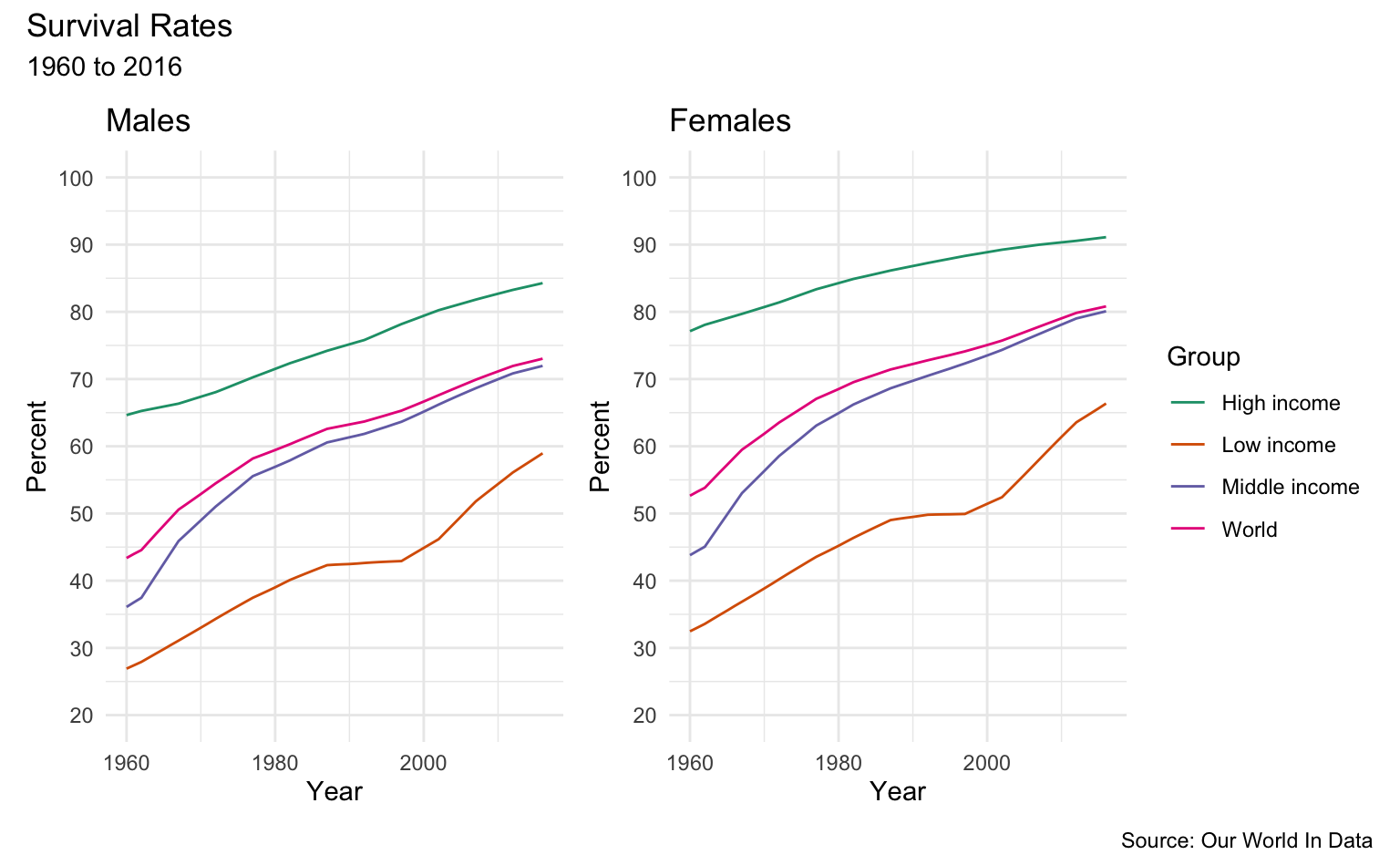
By Income Distribution Group World-wide
sample_list <- c("High income", "Middle income", "Low income", "World")
m <- men_survival_to_age_65 %>%
filter(Entity %in% sample_list) %>%
sr_plot("Males", "Group")
w <- women_survival_to_age_65 %>%
filter(Entity %in% sample_list) %>%
sr_plot("Females", "Group")
m + w + plot_layout(ncol = 2, guides = "collect") +
plot_annotation(
title = "Survival Rates",
subtitle = "1960 to 2016",
caption = "Source: Our World In Data"
)
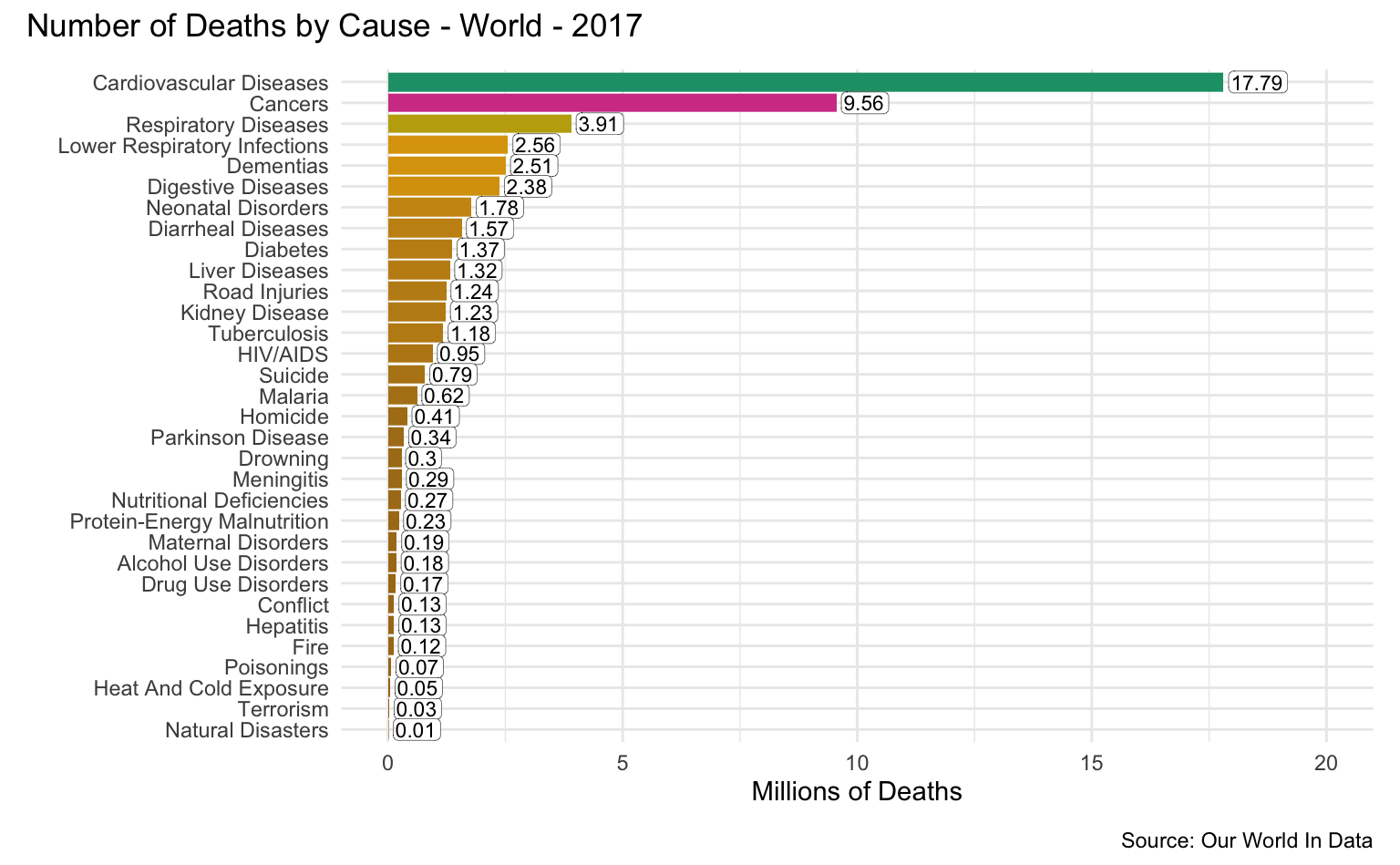
Causes of Death
The Global Burden of Disease is a major global study on the causes of death and disease published in the medical journal The Lancet. These estimates of the annual number of deaths by cause are shown in these graphics.
World
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "World") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Millions of Deaths", 20, 5)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - World - ", latest_year),
caption = "Source: Our World In Data"
)
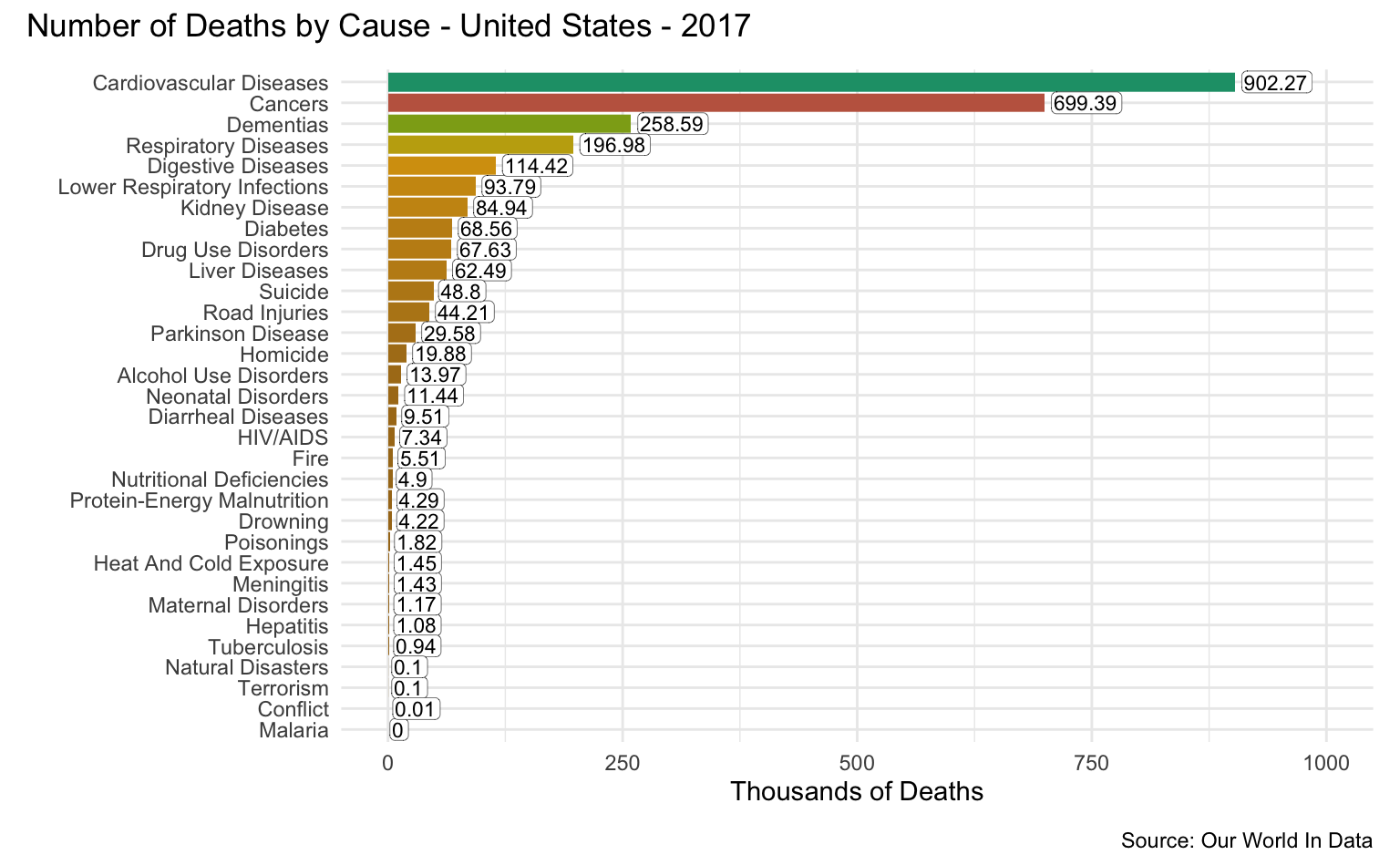
United States
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "United States") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Thousands of Deaths", 1000, 250)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - United States - ", latest_year),
caption = "Source: Our World In Data"
)
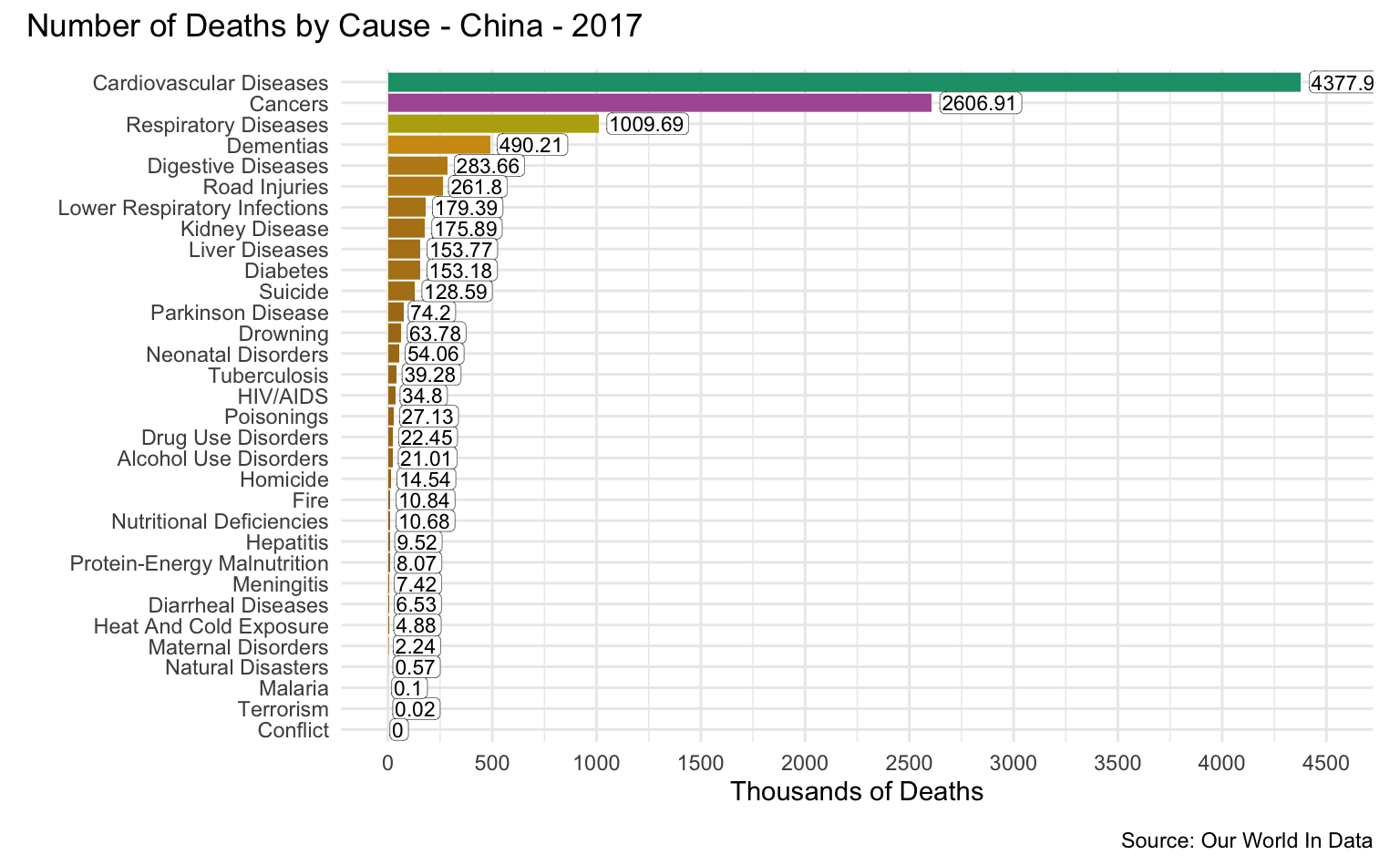
China
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "China") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Thousands of Deaths", 4500, 500)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - China - ", latest_year),
caption = "Source: Our World In Data"
)
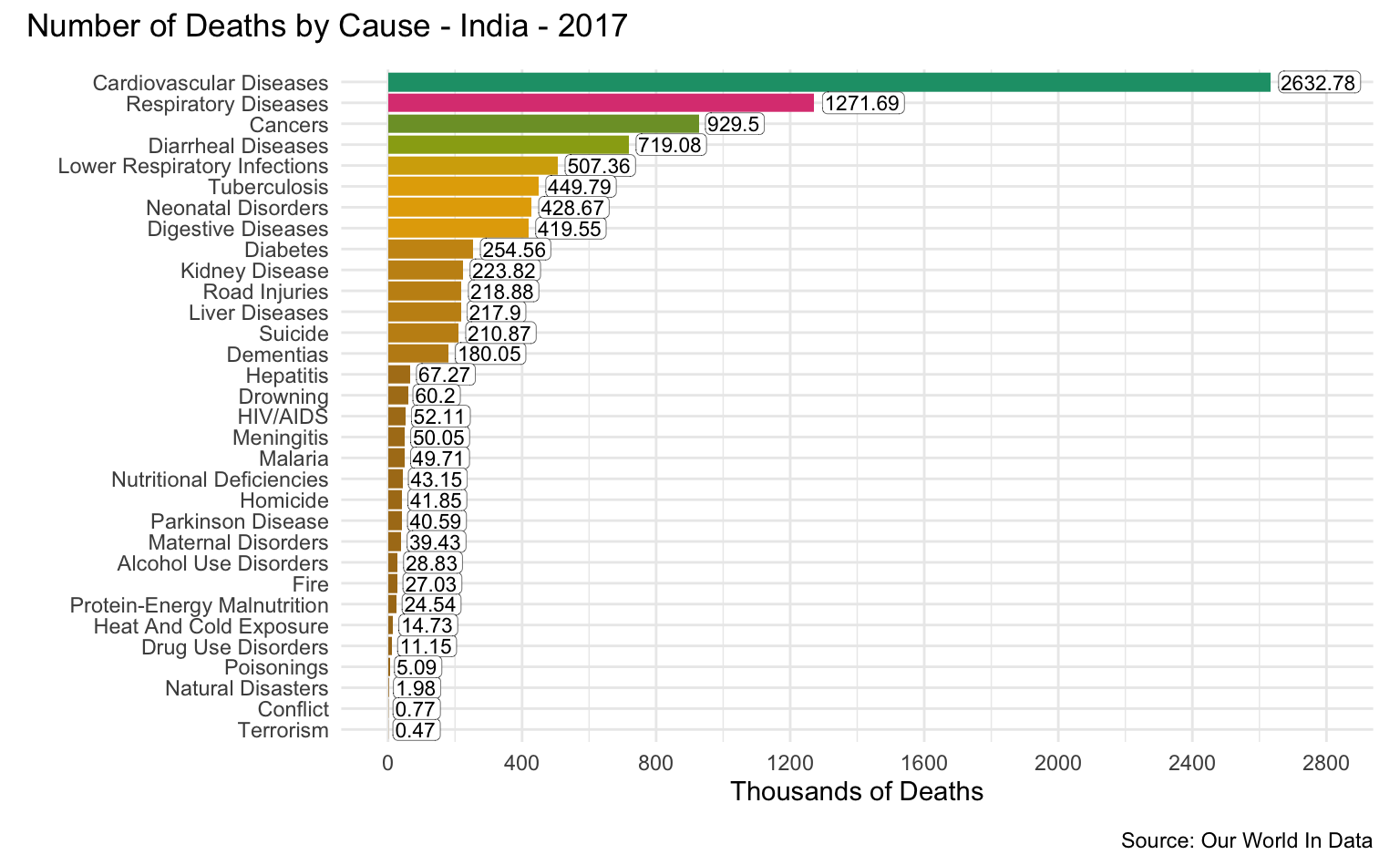
India
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "India") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Thousands of Deaths", 2800, 400)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - India - ", latest_year),
caption = "Source: Our World In Data"
)
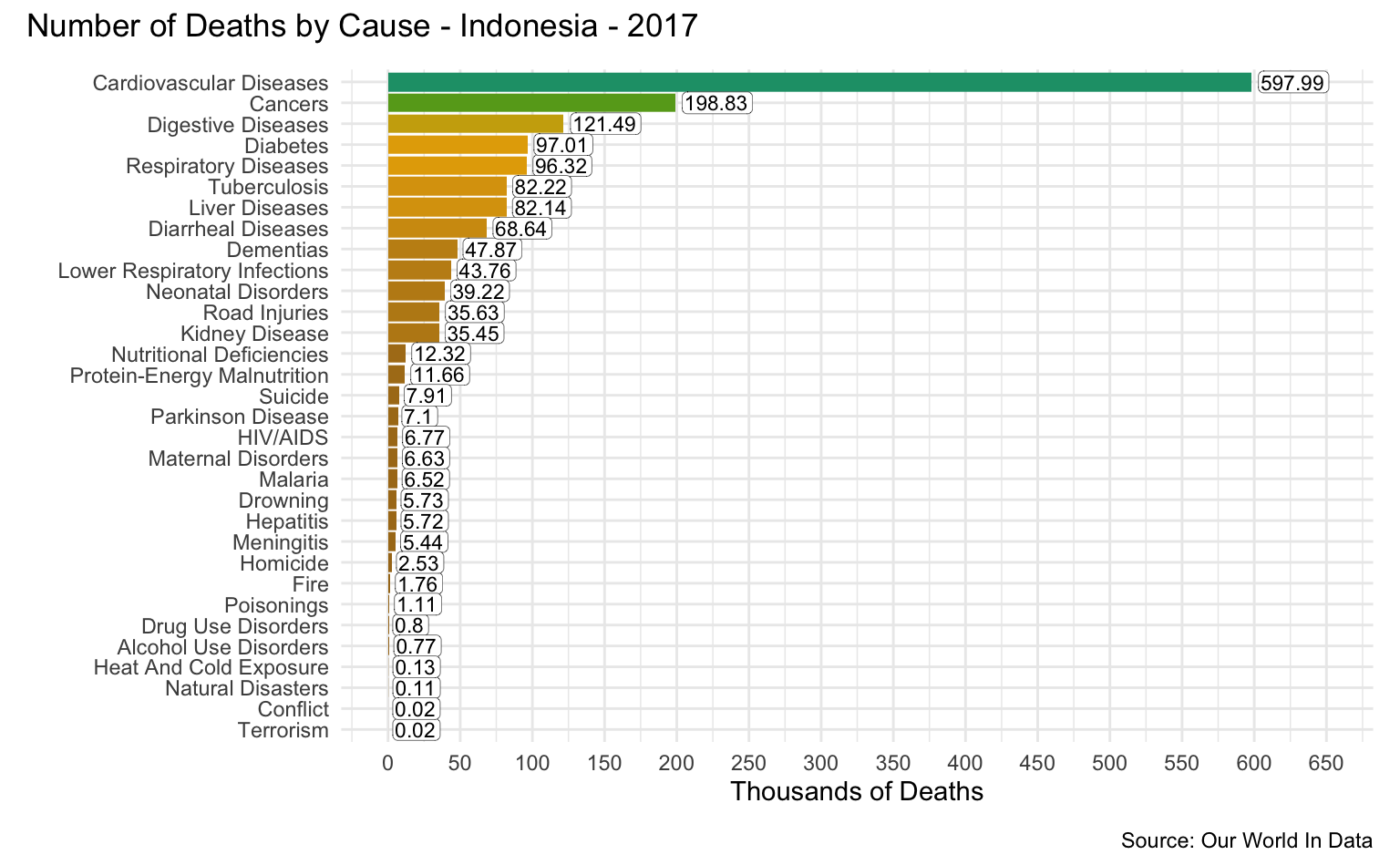
Indonesia
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "Indonesia") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Thousands of Deaths", 650, 50)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - Indonesia - ", latest_year),
caption = "Source: Our World In Data"
)
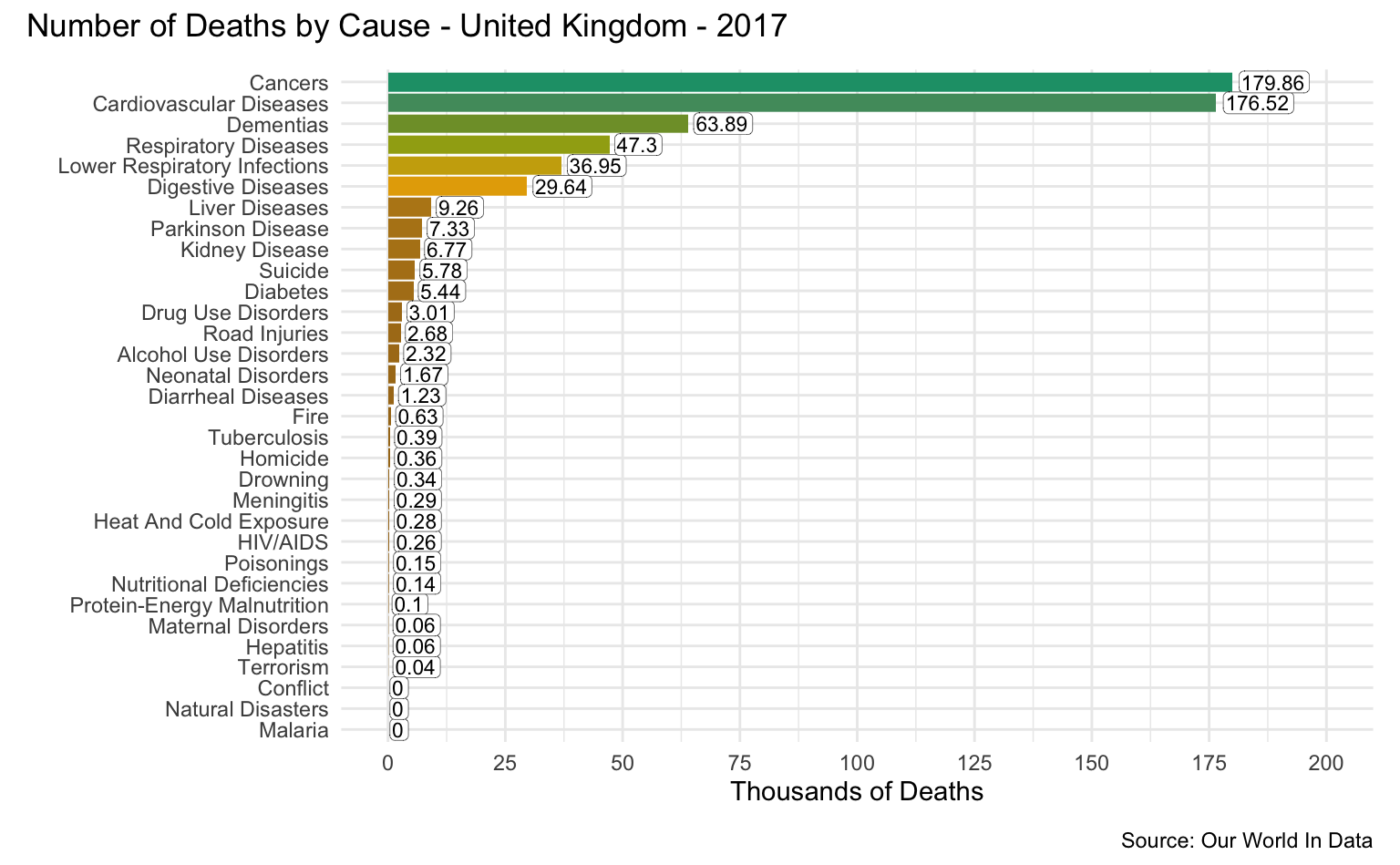
United Kingdom
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "United Kingdom") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Thousands of Deaths", 200, 25)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - United Kingdom - ", latest_year),
caption = "Source: Our World In Data"
)
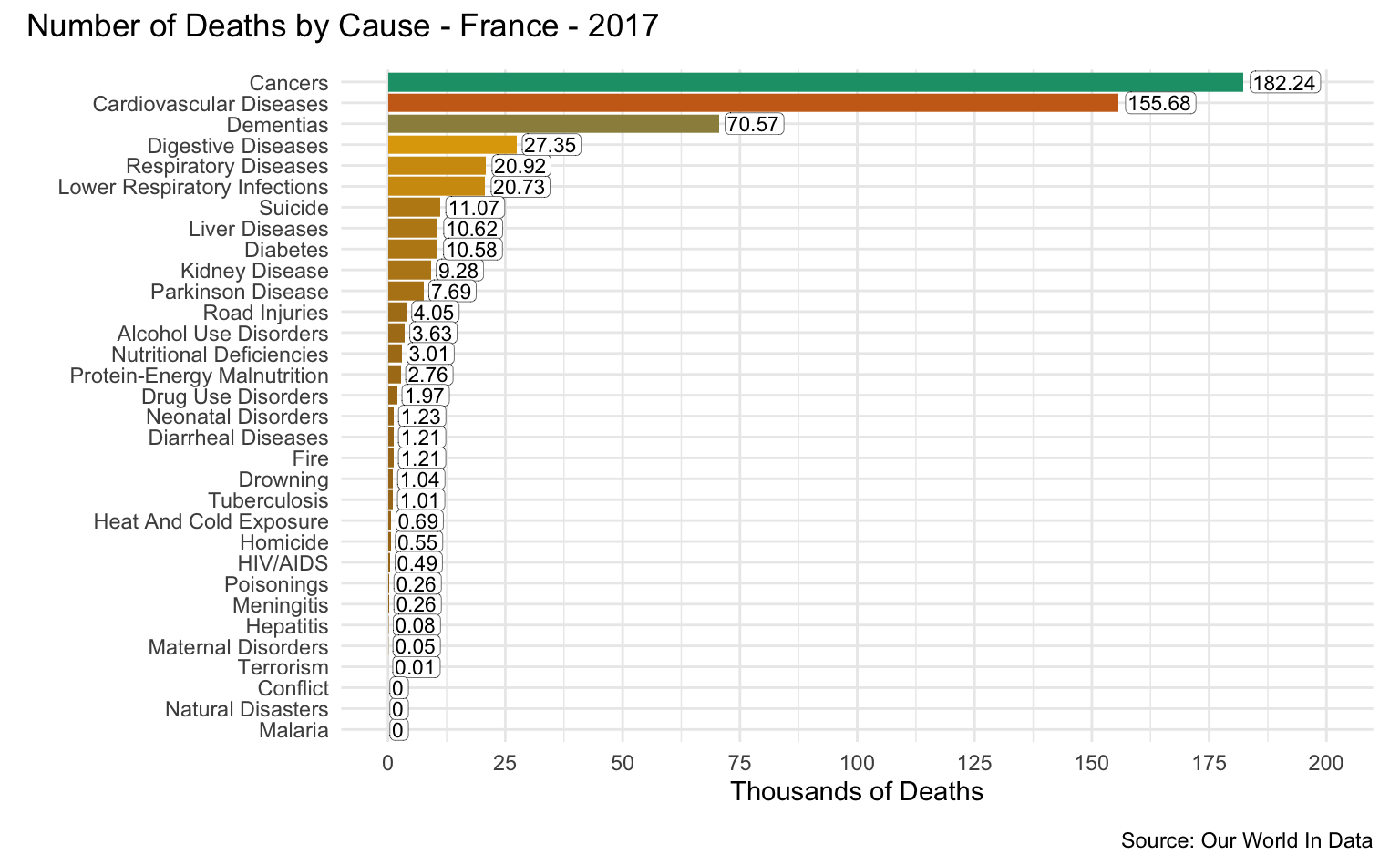
France
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "France") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Thousands of Deaths", 200, 25)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - France - ", latest_year),
caption = "Source: Our World In Data"
)
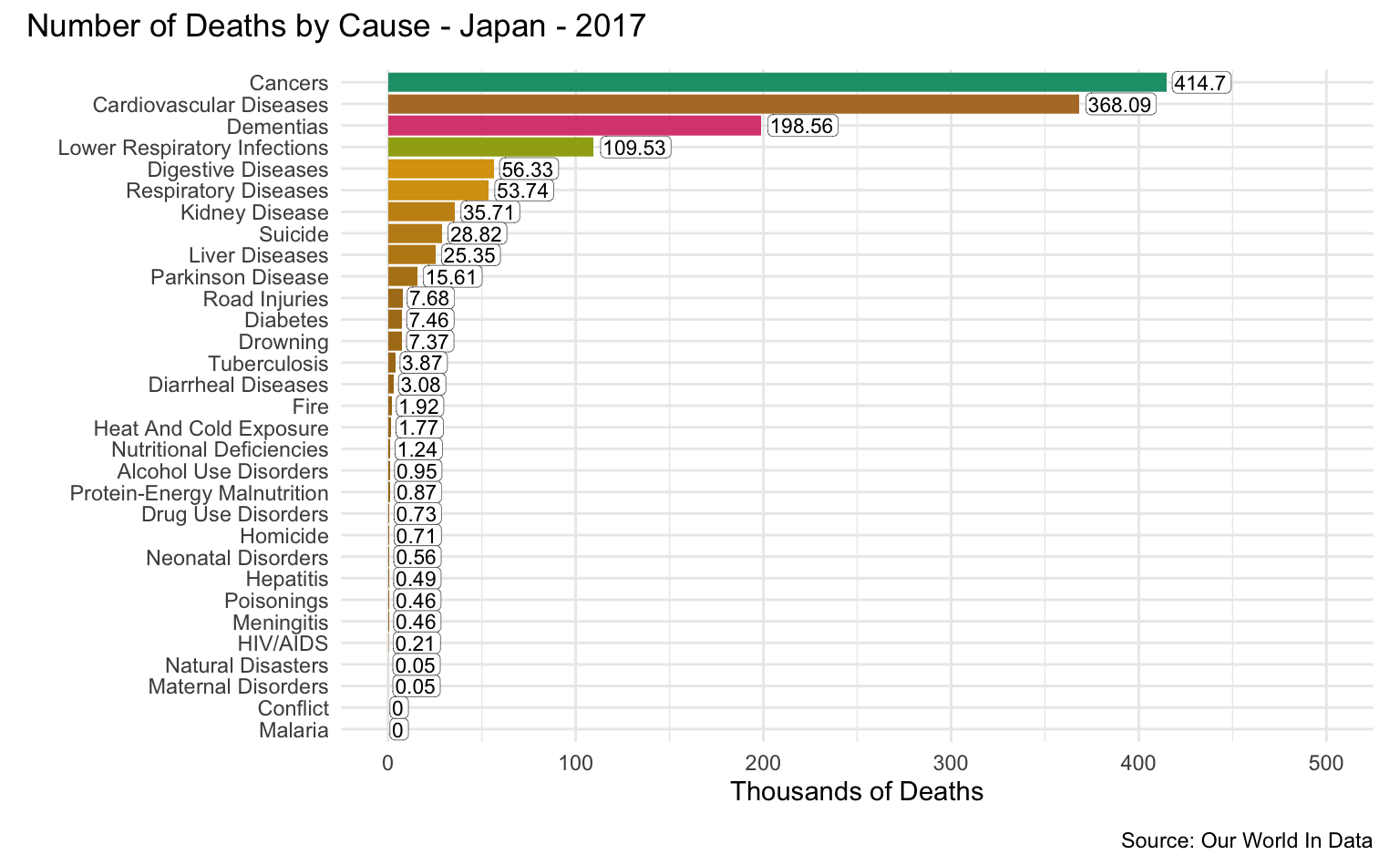
Japan
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "Japan") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Thousands of Deaths", 500, 100)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - Japan - ", latest_year),
caption = "Source: Our World In Data"
)
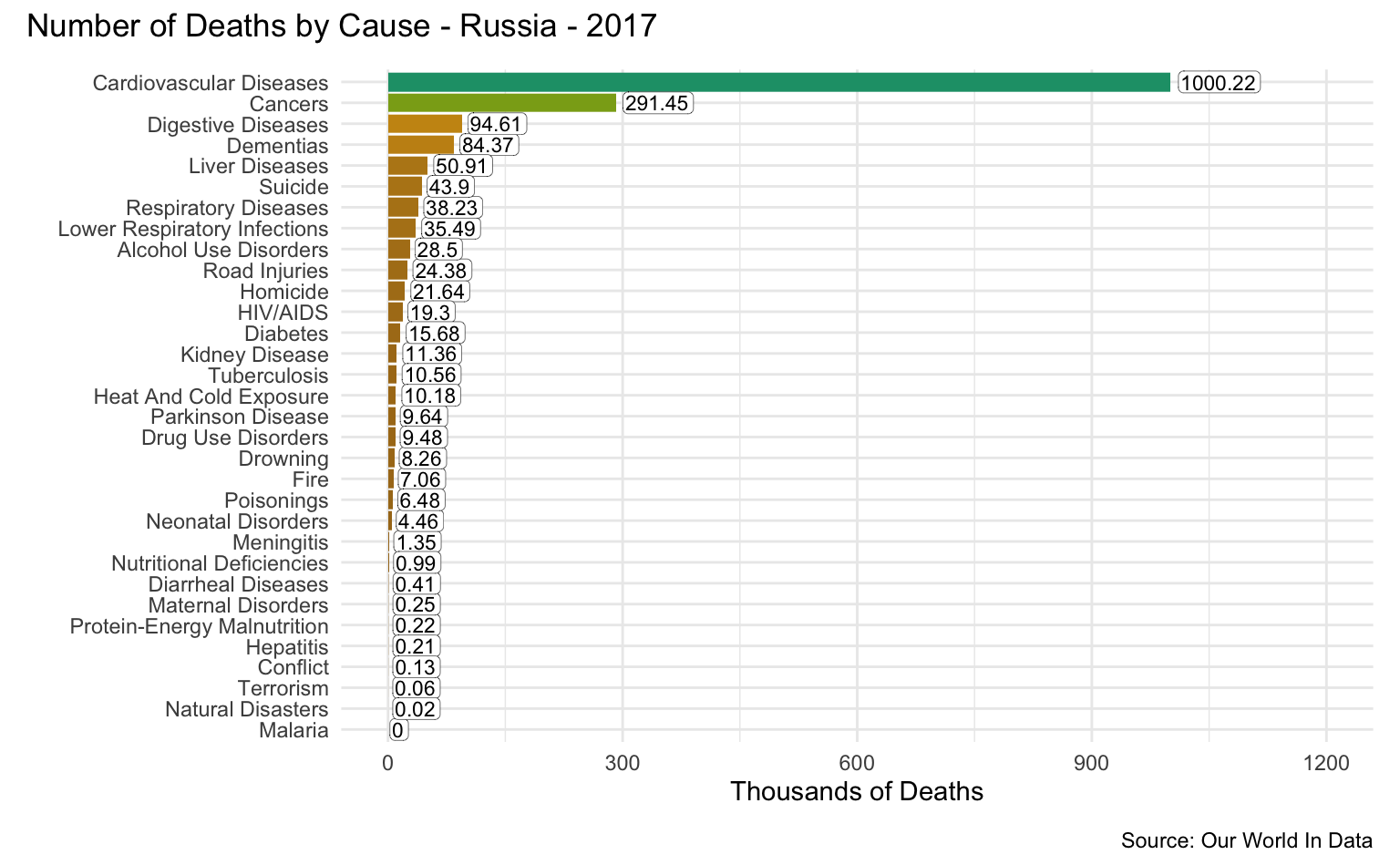
Russia
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "Russia") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Thousands of Deaths", 1200, 300)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - Russia - ", latest_year),
caption = "Source: Our World In Data"
)
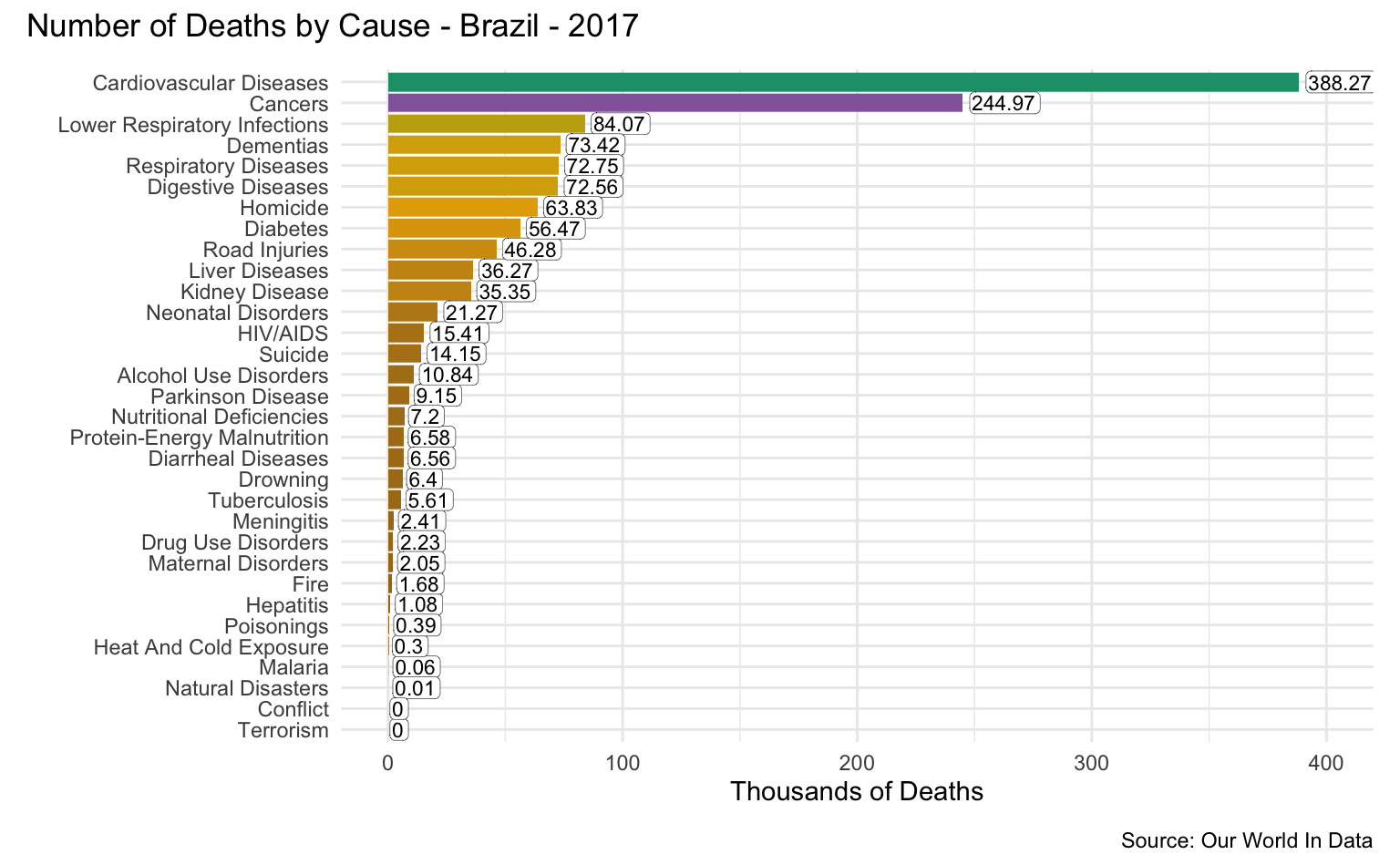
Brazil
latest_year <- max(causes_of_death$Year)
g <- causes_of_death %>%
filter(Entity == "Brazil") %>%
filter(Year == latest_year) %>%
pivot_longer(all_of(causes), names_to = "Cause", values_to = "Count") %>%
filter(!is.na(Count)) %>%
mutate(
Count = round(Count / 1000, 2),
Cause = reorder(as_factor(str_replace_all(Cause, "_", " ")), Count)
) %>%
cod_plot("Thousands of Deaths", 400, 100)
g + plot_annotation(
title = str_c("Number of Deaths by Cause - Brazil - ", latest_year),
caption = "Source: Our World In Data"
)